Computer science is not just the writing of programs or the development of code, it is the science of computation. While the field of computer science began with Alan Turing and John von Neumann in the 1940's, the origins of computation can be traced to the development of life on Earth. This view, that evolution and the resultant biological systems is in fact complex information processing, is helping scientists unlock the secrets of life in the emerging fields of systems and executable biology. Systems biology seeks to move beyond the classic reductionist approach and understand the biological world in terms of not just its component parts, but also the myriad interactions among these parts. Executable biology integrates the data and models of systems biology into computational tools that simulate the activity of living systems and facilitate "in silico" experiments leading to scientific and medical breakthroughs.
The field of systems biology has been successful in understanding cell components and their complex interactions through the integration of high throughput data sources with computational analysis. The challenge is to extend systems biology over multiple scales to comprehend how subcellular processes control cell behavior and in turn, how interactions among cells lead to large scale organization at the tissue level. Such knowledge is key to unlocking the genetic foundations of morphological development and disease.
Dr. Flann's research interests lie advancing executable biology by developing mechanistic multiscale models that bridge the gap between regulatory network dynamics and morphological outcomes. The work focuses on applying high-fidelity methods that implement the diversity of cell physiology, not directly as high level descriptions, but as combinations of modular subcellular mechanisms. One such modeling approach is the Cellular Potts Model (CPM) that represents 2D and 3D cellular systems as lattices of simple mesoscopic particles and model components as additive energy terms over cell and sub-cell configurations. The advantage for multiscale modeling is in its simplicity and realism since, just as in living systems, organization at the cell, multicell and tissue scale emerges through the complex interaction of lower-level mechanisms.
Areas of Research:
Research in Flann's lab is directed to the development and application of multiscale models to significant biological subsystems in development, cancer, immunity and yeast colony development. Through active collaboration with Vanderbilt, UCL and multiple labs at the Institute for Systems Biology in Seattle, common application-independent methodologies are being developed and applied to these specific domains as pilots systems. Some of the questions driving the research are:
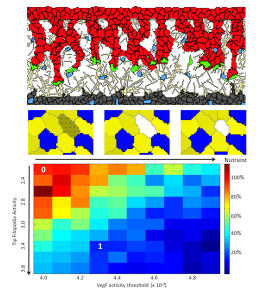
- What are the impacts of integrating models of intracellular regulatory networks into the CPM? This research seeks to understand how the temporal dynamics of regulatory networks at the subcellular scale influence the multi-cell spatiotemporal dynamics of morphology development. By linking regulation to morphology, the influence of small molecule interventions on tissue level manifestations of disease can be predicted and potential treatments discovered through high-throughput simulations. Previous work has demonstrated the feasibility of this approach in discovering potential subcellular interventions in angiogenesis that lead to disruptions in the organization of the vessel network and subsequent nutrient delivery to micro-tumors.
- How do the network dynamics and the attractor landscape of regulatory networks lead developmental systems to convergence to robust attractors in morphological space? Study of multiscale network dynamics aims to expand the established body of work in criticality of regulatory networks to include morphodynamic feedback among mechanisms such as cell/cell signaling and cell motility, apoptosis and proliferation. With such an extension, the tools of complexity could be applied to large-scale dynamic systems in order to recognize criticality in robust development and chaos in tissue level diseases such as cancer.
- How can multiscale experimental data directly inform and validate the models? Data sources span scales from regulatory networks induced from RNA-seq, microfluidic cytometry, multicell in vitro time-lapse images, to colorimetric markers that report spatial and temporal patterns of RNA expression over developing tissues. While methods exist for analyzing and validating data when viewed individually, methods are needed that link data sources over multiple scales so that data at one level can constrain interpretations of data at another level. Methods are under development to address this problem that work by identifying suggested model corrections as discrepancies between simulated and actual outcomes at one scale, and then perform model-based error propagation to other scales.
- How can high performance and cloud computing technology enable high-throughput multiscale model executions over large complex configurations of thousands of cells? As models incorporate more subcellular detail, cell/cell interactions and progress from the multicell scale to whole tissues, computational resources become a limiting bottleneck. Previous work has proved the value of massively parallel grid computing for model space exploration, but utilization of parallelism within individual simulations is an open problem. Collaboration between ISB, USU and Pacific Northwest National Laboratory (PNNL) high performance computing group is underway to develop effective solutions.
Key collaborations:
Gregory Podgorski (Utah State University): Projects include: (a) a multidisciplinary study in understanding combinations of developmental mechanisms and their interaction can produce regular mozaic patterns in the inner ear, which is critical to effective hearing. This project is in collaboration with Dr. Nicolas Daudent from the Ear Institute, University College London, who is working to understand the causes and potential cures of deafness; (b) utilizing multiscale models of angiogenesis to discover potential novel interventions to slow the growth of micro-tumors.
Ilya Shmulevich (Institute of Systems Biology): Projects include: (a) a multidisciplinary study of how glioma development is influenced by the interactions among the immune, vascular and micro-tumor systems. This work is in collaboration with Dr. Wei Zhang at MD Anderson Cancer Center and involves the integrated of in vitro experimentation, image analysis and multiscale modeling; (b) understanding criticality at multiple scales in morphological and disease development; and (c) the designing of new methods for model fitting and validation from multiscale images.
Ażmee Dudley (Institute of Systems Biology): Projects include: (a) the development of computational frameworks that tightly integrate whole yeast colony simulation with high-throughput experimentation; and (b) the application of multiscale models to understand sub-colony structure formation including carbohydrate matrix and its role in the development of complex and distinct colony features such as ridges, folds and aerial tubes.
Adrian Ozinsky (Institute of Systems Biology): Projects include model-based cell tracking from in vitro time-lapse images of epithelial to mesenchymal cell transition, which is a key step in the matastasis of breast tumors. The goal is to understand mechanisms of transition and discover possible precursor morphology and physiology clues that predict cell state changes.
Colette Calmelet (California State University, Chico): Projects include building models to understand the morphological development of the zebrafish notochord---a common feature of all chordates including humans---that is associated with congenitial disorders of the spinal cord. The project is in collaboration with an exprimental biologist Diane Sepich at Vanderbilt University.